January 2024
·
118 Reads
·
1 Citation
Microbiology Spectrum
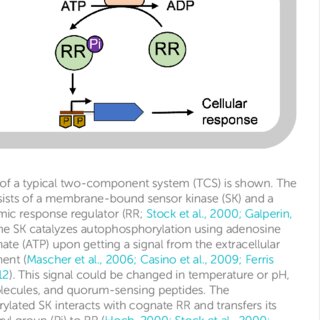
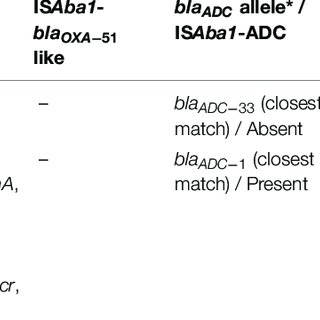
Colistin resistance in Acinetobacter baumannii is mediated by multiple mechanisms. Recently, mutations within pmrABC two-component system and overexpression of eptA gene due to upstream insertion of IS Aba1 have been shown to play a major role. Thus, the aim of our study is to characterize colistin resistance mechanisms among the clinical isolates of A. baumannii in India. A total of 207 clinical isolates of A. baumannii collected from 2016 to 2019 were included in this study. Mutations within lipid A biosynthesis and pmrABC genes were characterized by whole-genome shotgun sequencing. Twenty-eight complete genomes were further characterized by hybrid assembly approach to study insertional inactivation of lpx genes and the association of IS Aba1-eptA . Several single point mutations (SNPs), like M12I in pmrA , A138T and A444V in pmrB , and E117K in lpxD, were identified. We are the first to report two novel SNPs (T7I and V383I) in the pmrC gene. Among the five colistin-resistant A. baumannii isolates where complete genome was available, the analysis showed that three of the five isolates had IS Aba1 insertion upstream of eptA . No mcr genes were identified among the isolates. We mapped the SNPs on the respective protein structures to understand the effect on the protein activity. We found that majority of the SNPs had little effect on the putative protein function; however, some SNPs might destabilize the local structure. Our study highlights the diversity of colistin resistance mechanisms occurring in A. baumannii, and IS Aba1 -driven eptA overexpression is responsible for colistin resistance among the Indian isolates. IMPORTANCE Acinetobacter baumannii is a Gram-negative, emerging and opportunistic bacterial pathogen that is often associated with a wide range of nosocomial infections. The treatment of these infections is hindered by increase in the occurrence of A. baumannii strains that are resistant to most of the existing antibiotics. The current drug of choice to treat the infection caused by A. baumannii is colistin, but unfortunately, the bacteria started to show resistance to the last-resort antibiotic. The loss of lipopolysaccharides and mutations in lipid A biosynthesis genes are the main reasons for the colistin resistance. The present study characterized 207 A. baumannii clinical isolates and constructed complete genomes of 28 isolates to recognize the mechanisms of colistin resistance. We showed the mutations in the colistin-resistant variants within genes essential for lipid A biosynthesis and that cause these isolates to lose the ability to produce lipopolysaccharides.