May 2024
·
34 Reads
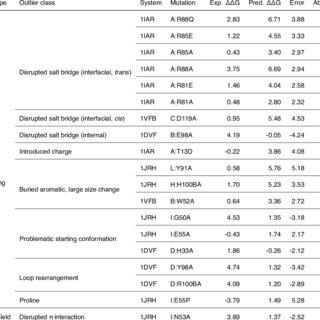
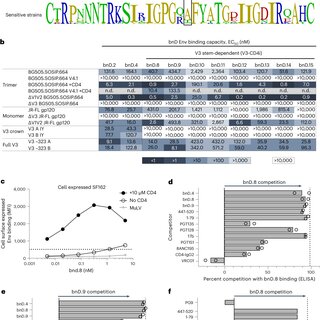
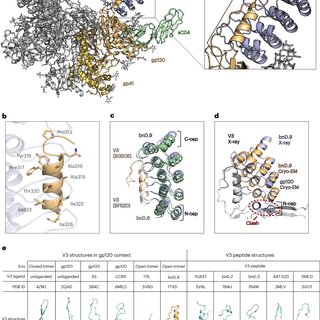
Broadly neutralizing antibodies are proposed as therapeutic and prophylactic agents against HIV‐1, but their potency and breadth are less than optimal. This study describes the immunization of a llama with the prefusion‐stabilized HIV‐1 envelope (Env) trimer, BG505 DS‐SOSIP, and the identification and improvement of potent neutralizing nanobodies recognizing the CD4‐binding site (CD4bs) of vulnerability. Two of the vaccine‐elicited CD4bs‐targeting nanobodies, G36 and R27, when engineered into a triple tandem format with llama IgG2a‐hinge region and human IgG1‐constant region (G36×3‐IgG2a and R27×3‐IgG2a), neutralized 96% of a multiclade 208‐strain panel at geometric mean IC80s of 0.314 and 0.033 µg mL⁻¹, respectively. Cryo‐EM structures of these nanobodies in complex with Env trimer revealed the two nanobodies to neutralize HIV‐1 by mimicking the recognition of the CD4 receptor. To enhance their neutralizing potency and breadth, nanobodies are linked to the light chain of the V2‐apex‐targeting broadly neutralizing antibody, CAP256V2LS. The resultant human‐llama bispecific antibody CAP256L‐R27×3LS exhibited ultrapotent neutralization and breadth exceeding other published HIV‐1 broadly neutralizing antibodies, with pharmacokinetics determined in FcRn‐Fc mice similar to the parent CAP256V2LS. Vaccine‐elicited llama nanobodies, when combined with V2‐apex broadly neutralizing antibodies, may therefore be able to fulfill anti‐HIV‐1 therapeutic and prophylactic clinical goals.