A preview of this full-text is provided by American Society for Microbiology.
Content available from Journal of Clinical Microbiology
This content is subject to copyright. Terms and conditions apply.
JOURNAL OF CLINICAL MICROBIOLOGY, July 2010, p. 2563–2564 Vol. 48, No. 7
0095-1137/10/$12.00 doi:10.1128/JCM.01905-09
Copyright © 2010, American Society for Microbiology. All Rights Reserved.
Multilocus Sequence Types of Carbapenem-Resistant
Pseudomonas aeruginosa in Singapore Carrying
Metallo--Lactamase Genes, Including
the Novel bla
IMP-26
Gene
䌤
Tse Hsien Koh,*
1
Cheng Teng Khoo,
1
Thuan Tong Tan,
2
Mohamed Amir Bin Mohamed Arshad,
3
Li Ping Ang,
3
Lee Jin Lau,
3
Li-Yang Hsu,
4
and Eng Eong Ooi
5
Department of Pathology, Singapore General Hospital, Outram Road, 169608 Singapore
1
; Department of Infectious Diseases,
Singapore General Hospital, Outram Road, 169608 Singapore
2
; Temasek Applied Science School, Temasek Polytechnic,
21 Tamplines Avenue 1, 529757 Singapore
3
; Department of Medicine, Yong Loo Lin School of Medicine, National University of
Singapore, 5 Lower Kent Ridge Road, 119074 Singapore
4
; and Duke-NUS Graduate Medical School,
8 College Road, 169857 Singapore
5
Received 26 September 2009/Returned for modification 13 October 2009/Accepted 26 April 2010
Nine imipenem-resistant Pseudomonas aeruginosa isolates were found to contain a variety of metallo--
lactamase genes, including bla
IMP-1
,bla
IMP-7
,bla
VIM-2
,bla
VIM-6
, and the novel bla
IMP-26
. Multilocus sequence
typing showed a diversity of sequence types. Comparison with isolates from an earlier study showed that the
epidemic clones from 2000 have not become established.
Carbapenem-resistant Pseudomonas aeruginosa is an in-
creasing problem worldwide. While many underlying mecha-
nisms may account for carbapenem resistance in this species,
the possession of metallo--lactamase (MBL) genes is of par-
ticular concern because these enzymes are able to hydrolyze all
-lactam antimicrobials with the exception of aztreonam. In
addition, these genes may be mobilized and transferred be-
tween different species of bacteria. We conducted a study in
2008 to investigate if there were any changes in the epidemi-
ology of P. aeruginosa isolates containing MBL genes in our
hospital compared to results from an earlier survey carried out
in 2000 (3).
Of 2,552 nonduplicate P. aeruginosa organisms isolated in
2008, 123 isolates were imipenem resistant. Of these, 11 were
positive for MBL production by imipenem-EDTA disk diffu-
sion (5). Nine of these yielded a product by multiplex PCR for
MBL genes (2). The individual MBL genes were then ampli-
fied and sequenced. The clonal relationship between isolates
with MBL genes was determined by pulsed-field gel electro-
phoresis (PFGE) of chromosomal DNA restricted with SpeI
(3). The PFGE band patterns were analyzed with Bionumerics
(Applied Maths NV, Sint-Martens-Latem, Belgium), and all
strains with more than 85% similarity were considered to be-
long to the same clone. All strains were further subjected to
multilocus sequence typing (MLST) (1). Because it is a nucleic
acid sequence-based method, MLST is able to characterize
bacterial types in an unambiguous fashion and establish evo-
lutionary relationships between strains better than band-based
methods like PFGE. Representative MBL-producing P. aerugi-
nosa isolates from the 2000 survey were also subjected to
PFGE and MLST. MLST profiles were submitted to eBURST
V3 (http://eburst.mlst.net/) on 10 March 2010. Isolates sharing
six out of seven alleles were assigned to the same BURST
group and can be considered to belong to the same clonal
complex descended from a common founder genotype. The
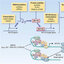
PFGE, MBL gene sequence, and MLST results are summa-
rized in Fig. 1.
In our previous study, 21 of 2,094 (1.0%) of all nonduplicate
P. aeruginosa isolates in our hospital had MBL genes (3). With
the exception of one isolate with bla
IMP-7
, all other isolates had
bla
IMP-1
and belonged to one of two PFGE clones. Isolates
belonging to clone A had sequences identical to that of the
original bla
IMP-1
first reported in Japan. Four representatives
of clone A isolated from our hospital in 2000 had sequence
type 964 (ST964) by MLST. Isolates belonging to clone B
isolated in 2000 had sequences for variant bla
IMP-1
(bla
IMP-1v
)
with four silent mutations. Three representatives of this clone
from 2000 had ST233 and one had ST742 based on MLST. All
four representatives of clone B belong to the same BURST
group, which was different from that of clone A.
In contrast, in the 2008 survey, 9 of 2,552 (0.35%) nondu-
plicate P. aeruginosa isolates had MBL genes. Unlike the ear-
lier study, there were no large clonal outbreaks. Two isolates
with bla
IMP-1v
had similar PFGE patterns and belonged to the
same BURST group as representative isolates from clone B in
2000.
Two isolates from 2008 with bla
IMP-7
had similar PFGE
patterns and shared the same BURST group. The rest of the
isolates from 2008 had distinct PFGE patterns.
There was a greater diversity of MBL genes compared to the
2000 survey results. In particular, this is the first time that
bla
VIM-2
and bla
VIM-6
have been found in P. aeruginosa in
Singapore. bla
IMP-26
is a novel MBL gene that differs from
bla
IMP-4
at position 145 (G-to-T change). The translated amino
acid sequence differs from IMP-4 at residue 49 (phenylalanine
* Corresponding author. Mailing address: Department of Pathology,
Singapore General Hospital, Outram Road, 169608 Singapore. Phone:
65-63214275. Fax: 65-62226826. E-mail: koh.tse.hsien@sgh.com.sg.
䌤
Published ahead of print on 12 May 2010.
2563
for valine). This sequence has been previously deposited in the
GenBank database as IMP-4 from an Acinetobacter calcoace-
ticus isolate from Malaysia (accession number ABC24668.1).
Three of the isolates in this study (separately containing
bla
VIM-2
,bla
IMP-1
, and bla
IMP-7
) belonged to ST235. This se-
quence type has been described in a VIM-producing P. aerugi-
nosa isolate in Belgrade and is the founder of an international
clonal complex of isolates bearing MBL genes found in several
countries in Europe (6). Recently, an increasing prevalence of
IMP-1-producing P. aeruginosa has been found in Hiroshima,
Japan. This was due entirely to the clonal expansion of only
two lineages, ST235 (BURST group 3) and ST357 (BURST
group 108) (4). This is similar to the situation that existed in
Singapore in 2000, where only two lineages (BURST groups 29
and 44) accounted for the majority of MBL-producing P.
aeruginosa (3).
It is noteworthy that the original fear that a clone of MBL-
producing P. aeruginosa would become established in Singa-
pore has not been realized. The BURST group 29 and 44
lineages from 2000 were represented by only one to two iso-
lates in 2008. The two P. aeruginosa isolates with bla
IMP-7
in
2008 are unrelated to the solitary isolate with bla
IMP-7
from
2000. It has been suggested that P. aeruginosa displays an
epidemic population structure, with a limited number of clones
emerging from a large number of unrelated genotypes (7).
Although we did not correlate our study with hospital infection
control measures, the Japanese data and our own seem to
suggest that controlling the prevalence of MBL-producing P.
aeruginosa may be achieved by preventing the transmission of
specific epidemic clones.
While it is reassuring to note that the prevalence of MBL
producers in carbapenem-resistant P. aeruginosa has not in-
creased, the increased diversity of MBL genes represents a new
cause for concern. We were unable to characterize the gene
responsible for the MBL phenotype in two isolates in this
study, and these may represent novel resistance determinants.
Although clones of MBL-producing P. aeruginosa have not
become established, it seems likely, given the variation of MBL
genes and MLST types in this study, that MBL-producing P.
aeruginosa continues to be introduced to our hospital from
diverse sources.
Nucleotide sequence accession number. The sequence for
bla
IMP-26
was submitted to GenBank under the accession num-
ber GU045307.
We acknowledge Ong Lan Huay for technical assistance.
REFERENCES
1. Curran, B., D. Jonas, H. Grundmann, T. Pitt, and C. G. Dowson. 2004.
Development of a multilocus sequence typing scheme for the opportunistic
pathogen Pseudomonas aeruginosa. J. Clin. Microbiol. 42:5644–5649.
2. Ellington, M. J., J. Kistler, D. M. Livermore, and N. Woodford. 2007. Mul-
tiplex PCR for rapid detection of genes encoding acquired metallo--lacta-
mases. J. Antimicrob. Chemother. 59:321–322.
3. Koh, T. H., G. C. Wang, and L. H. Sng. 2004. Clonal spread of IMP-1-
producing Pseudomonas aeruginosa in two hospitals in Singapore. J. Clin.
Microbiol. 42:5378–5380.
4. Kouda, S., M. Ohara, M. Onodera, Y. Fujiue, M. Sasaki, T. Kohara, S.
Kashiyama, S. Hayashida, T. Harino, T. Tsuji, H. Itaha, N. Gotoh, A. Mat-
subara, T. Usui, and M. Sugai. 2009. Increased prevalence and clonal dis-
semination of multidrug-resistant Pseudomonas aeruginosa with the bla
IMP-1
gene cassette in Hiroshima. J. Antimicrob. Chemother. 64:46–51.
5. Lee, K., Y. Chong, H. B. Shin, Y. A. Kim, D. Yong, and J. H. Yum. 2001.
Modified Hodge and EDTA-disk synergy tests to screen metallo--lactamase-
producing strains of Pseudomonas and Acinetobacter species. Clin. Microbiol.
Infect. 7:88–91.
6. Lepsanovic, Z., B. Libisch, B. Tomanovic, Z. Nonkovici, B. Balogh, and M.
Fuzi. 2008. Characterisation of the first VIM metallo--lactamase-producing
Pseudomonas aeruginosa clinical isolate in Serbia. Acta Microbiol. Immunol.
Hung. 55:447–454.
7. Pirnay, J. P., D. De Vos, C. Cochez, F. Bilocq, A. Vanderkelen, M. Zizi, B.
Ghysels, and P. Cornelis. 2002. Pseudomonas aeruginosa displays an epidemic
population structure. Environ. Microbiol. 4:898–911.
FIG. 1. Dendrogram of PFGE patterns of P. aeruginosa isolates with metallo--lactamase genes, showing the year of isolation, MLST sequence
type, and BURST group.
2564 NOTES J. CLIN.MICROBIOL.
Content uploaded by Thean Yen Tan
Author content
All content in this area was uploaded by Thean Yen Tan
Content may be subject to copyright.